tick.survival¶
This module provides some tools for survival analysis. It provides a learner for Cox regression and a model for Self Control Case Series (SCCS).
1. Learners¶
For now, this module provides only a learner for Cox regression for proportional hazards, using the partial likelihood. It also provides Nelson-Aalen and Kaplan-Meier estimators.
|
Cox regression learner, using the partial Cox likelihood for proportional risks, with many choices of penalization. |
|
Computes the Nelson-Aalen cumulative hazard rate estimation given by: |
|
Computes the Kaplan-Meier survival function estimation given by: |
|
ConvSCCS learner, estimates lagged features effect using TV and Group L1 penalties. |
|
BatchConvSCCS provides parallel batch solving for ConvSCCS k_fold |
|
StreamConvSCCS provides parallel solving for k_fold and bootstrap ConvSCCS |
2. Models¶
The following models can be used with solvers for inference. Note that
ModelCoxRegPartialLik cannot
be used for now with a stochastic solver, while we can with ModelSCCS.
Partial likelihood of the Cox regression model (proportional hazards). |
|
|
Discrete-time Self Control Case Series (SCCS) likelihood. |
3. Simulation¶
We provide tools for the simulation of datasets, for both right-censored
durations following a proportional risks model, and a SCCS model.
SimuCoxReg generates data in the form of i.i.d triplets \((x_i, t_i, c_i)\)
for \(i=1, \ldots, n\), where \(x_i \in \mathbb R^d\) is a features vector,
\(t_i \in \mathbb R_+\) is the survival time and \(c_i \in \{ 0, 1 \}\) is the
indicator of right censoring.
Note that \(c_i = 1\) means that \(t_i\) is a failure time
while \(c_i = 0\) means that \(t_i\) is a censoring time.
|
Simulation of a Cox regression for proportional hazards |
|
Simulation of a Self Control Case Series (SCCS) model. |
Example¶
"""
==============================
Cox regression data simulation
==============================
Generates Cox Regression realization given a weight vector
"""
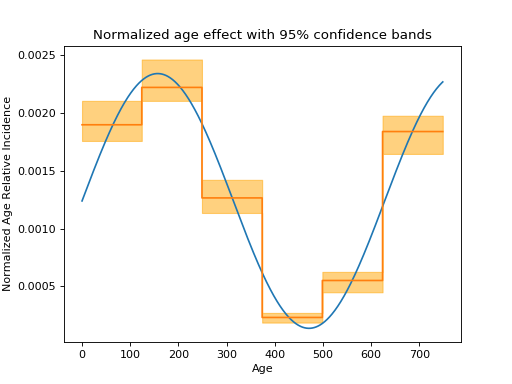
import matplotlib.pyplot as plt
import numpy as np
from tick.survival import SimuCoxReg
n_samples = 150
weights = np.array([0.3, 1.2])
seed = 123
simu_coxreg = SimuCoxReg(weights, n_samples=n_samples, seed=123, verbose=False)
X, T, C = simu_coxreg.simulate()
plt.figure(figsize=(6, 4))
plt.scatter(*X[C == 0].T, c=T[C == 0], cmap='RdBu', marker="x",
label="censoring")
plt.scatter(*X[C == 1].T, c=T[C == 1], cmap='RdBu', marker="o",
label="failure")
plt.colorbar()
plt.legend(loc='upper left')
plt.title('Cox regression', fontsize=16)
(Source code, png, hires.png, pdf)
"""
====================================================================
ConvSCCS cross validation on simulated longitudinal features example
====================================================================
In this example we simulate longitudinal data with preset relative incidence
for each feature. We then perform a cross validation of the ConvSCCS model
and compare the estimated coefficients to the relative incidences used for
the simulation.
"""
from time import time
import numpy as np
from scipy.sparse import csr_matrix, hstack
from matplotlib import cm
import matplotlib.pylab as plt
from tick.survival.simu_sccs import CustomEffects
from tick.survival import SimuSCCS, ConvSCCS
from mpl_toolkits.axes_grid1 import make_axes_locatable
# Simulation parameters
seed = 0
lags = 49
n_samples = 2000
n_intervals = 750
n_corr = 3
# Relative incidence functions used for the simulation
ce = CustomEffects(lags + 1)
null_effect = [ce.constant_effect(1)] * 2
intermediate_effect = ce.bell_shaped_effect(2, 30, 15, 15)
late_effects = ce.increasing_effect(2, curvature_type=4)
sim_effects = [*null_effect, intermediate_effect, late_effects]
n_features = len(sim_effects)
n_lags = np.repeat(lags, n_features).astype('uint64')
coeffs = [np.log(c) for c in sim_effects]
# Time drift (age effect) used for the simulations.
time_drift = lambda t: np.log(8 * np.sin(.01 * t) + 9)
# Simaltion of the features.
sim = SimuSCCS(n_samples, n_intervals, n_features, n_lags,
time_drift=time_drift, coeffs=coeffs, seed=seed,
n_correlations=n_corr, verbose=False)
features, censored_features, labels, censoring, coeffs = sim.simulate()
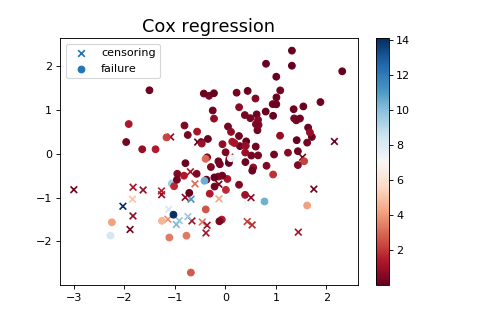
# Plot the Hawkes kernel matrix used to generate the features.
fig, ax = plt.subplots(figsize=(7, 6))
heatmap = ax.pcolor(sim.hawkes_exp_kernels.adjacency, cmap=cm.Blues)
divider = make_axes_locatable(ax)
cax = divider.append_axes("right", size="5%", pad=0.5)
fig.colorbar(heatmap, cax=cax)
ax.set_title('Hawkes adjacency matrix used for the simulation')
plt.show()
(Source code, png, hires.png, pdf)
## Add age_groups features to feature matrices.
agegrps = [0, 125, 250, 375, 500, 625, 750]
n_agegrps = len(agegrps) - 1
feat_agegrp = np.zeros((n_intervals, n_agegrps))
for i in range(n_agegrps):
feat_agegrp[agegrps[i]:agegrps[i + 1], i] = 1
feat_agegrp = csr_matrix(feat_agegrp)
features = [hstack([f, feat_agegrp]).tocsr() for f in features]
censored_features = [
hstack([f, feat_agegrp]).tocsr() for f in censored_features
]
n_lags = np.hstack([n_lags, np.zeros(n_agegrps)])
# Learning
# Example code for cross validation
# start = time()
# learner = ConvSCCS(n_lags=n_lags.astype('uint64'),
# penalized_features=np.arange(n_features),
# random_state=42)
# C_TV_range = (1, 4)
# C_L1_range = (2, 5)
# _, cv_track = learner.fit_kfold_cv(features, labels, censoring,
# C_TV_range, C_L1_range,
# confidence_intervals=True,
# n_samples_bootstrap=20, n_cv_iter=50)
# elapsed_time = time() - start
# print("Elapsed time (model training): %.2f seconds \n" % elapsed_time)
# print("Best model hyper parameters: \n")
# print("C_tv : %f \n" % cv_track.best_model['C_tv'])
# print("C_group_l1 : %f \n" % cv_track.best_model['C_group_l1'])
# cv_track.plot_cv_report(35, 45)
# plt.show()
# confidence_intervals = cv_track.best_model['confidence_intervals']
# using the parameters resulting from cross-validation
learner = ConvSCCS(
n_lags=n_lags.astype('uint64'), penalized_features=np.arange(n_features),
random_state=42, C_tv=270.2722840570933, C_group_l1=5216.472772625124)
_, confidence_intervals = learner.fit(features, labels, censoring,
confidence_intervals=True,
n_samples_bootstrap=20)
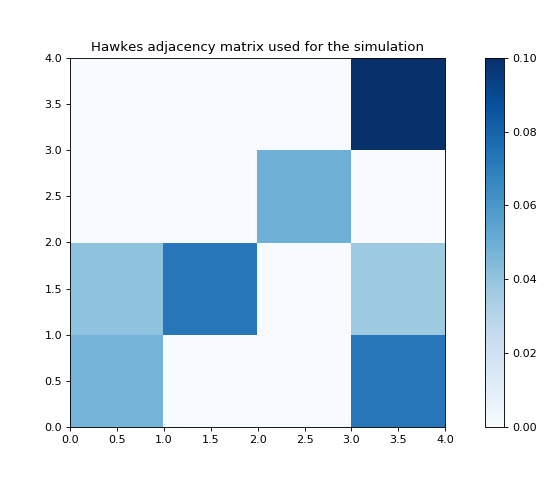
# Plot estimated parameters
# get bootstrap confidence intervals
refitted_coeffs = confidence_intervals['refit_coeffs']
lower_bound = confidence_intervals['lower_bound']
upper_bound = confidence_intervals['upper_bound']
n_rows = int(np.ceil(n_features / 2))
remove_last_plot = (n_features % 2 != 0)
fig, axarr = plt.subplots(n_rows, 2, sharex=True, sharey=True, figsize=(10, 6))
y = confidence_intervals['refit_coeffs']
lb = confidence_intervals['lower_bound']
ub = confidence_intervals['upper_bound']
for i, c in enumerate(y[:-6]):
ax = axarr[i // 2][i % 2]
l = n_lags[i]
ax.plot(np.exp(coeffs[i]), label="True RI")
ax.step(np.arange(l + 1), np.exp(c), label="Estimated RI")
ax.fill_between(
np.arange(l + 1), np.exp(lb[i]), np.exp(ub[i]), alpha=.5,
color='orange', step='pre', label="95% boostrap CI")
plt.suptitle('Estimated relative risks with 95% confidence bands')
axarr[0][1].legend(loc='best')
[ax[0].set_ylabel('Relative incidence') for ax in axarr]
[ax.set_xlabel('Time after exposure start') for ax in axarr[-1]]
if remove_last_plot:
fig.delaxes(axarr[-1][-1])
plt.show()
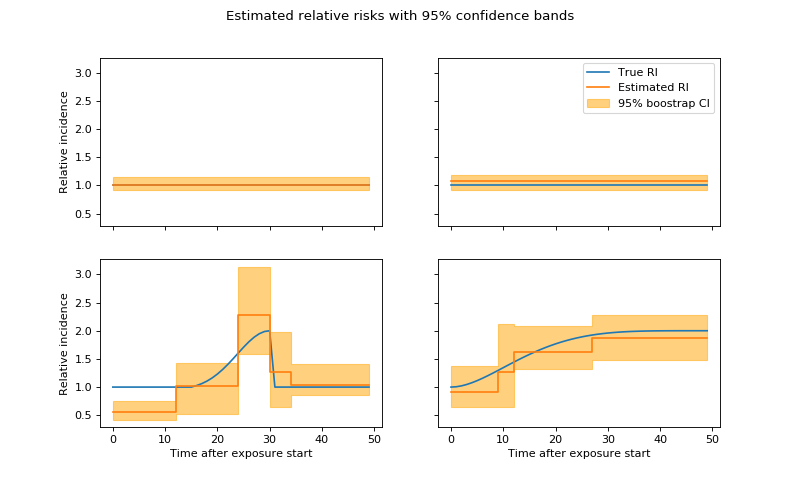
normalize = lambda x: x / np.sum(x)
m = np.repeat(np.hstack(refitted_coeffs[-6:]), 125)
lb = np.repeat(np.hstack(lower_bound[-6:]), 125)
ub = np.repeat(np.hstack(upper_bound[-6:]), 125)
plt.figure()
plt.plot(
np.arange(n_intervals),
normalize(np.exp(time_drift(np.arange(n_intervals)))))
plt.step(np.arange(n_intervals), normalize(np.exp(m)))
plt.fill_between(
np.arange(n_intervals),
np.exp(lb) / np.exp(m).sum(),
np.exp(ub) / np.exp(m).sum(), alpha=.5, color='orange', step='pre')
plt.xlabel('Age')
plt.ylabel('Normalized Age Relative Incidence')
plt.title("Normalized age effect with 95% confidence bands")
plt.show()