tick.linear_model¶
This module proposes tools for the inference and simulation of (generalized) linear models, including among others linear, logistic and Poisson regression, with a large set of penalization techniques and solvers. It also proposes hinge losses for supervised learning, namely the hinge, quadratic hinge and smoothed hinge losses.
1. Introduction¶
Given training data \((x_i, y_i) \in \mathbb R^d \times \mathbb R\)
for \(i=1, \ldots, n\), tick considers, when solving a generalized linear
model an objective of the form
where:
\(w \in \mathbb R^d\) is a vector containing the model weights;
\(b \in \mathbb R\) is the population intercept;
\(\ell : \mathbb R^2 \rightarrow \mathbb R\) is a loss function
\(g\) is a penalization function, see tick.prox for a list of available penalization technique
Depending on the model the label can be binary \(y_i \in \{ -1, 1 \}\) (binary classification), discrete \(y_i \in \mathbb N\) (Poisson models, see below) or continuous \(y_i \in \mathbb R\) (least-squares regression). The loss function \(\ell\) depends on the considered model, that are listed in 3. Models below. This modules proposes also end-user learner classes described below.
2. Learners¶
The following table lists the different learners that allows to train a model with several choices of penalizations and solvers.
Linear regression learner, with many choices of penalization and solvers. |
|
Logistic regression learner, with many choices of penalization and solvers. |
|
|
Poisson regression learner, with exponential link function. |
These classes follow whenever possible the scikit-learn API, namely fit
and predict methods, and follow the same naming conventions.
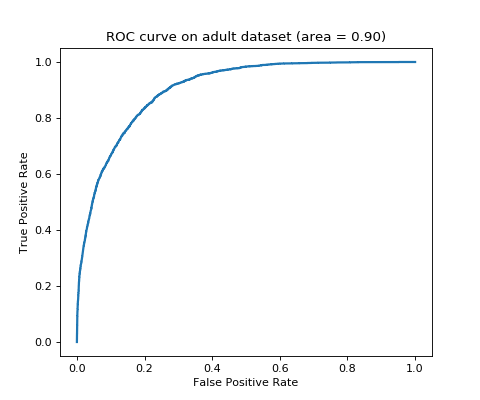
Example¶
"""
==============================================
Binary classification with logistic regression
==============================================
This code perform binary classification on adult dataset with logistic
regression learner (`tick.inference.LogisticRegression`).
"""
import matplotlib.pyplot as plt
from sklearn.metrics import roc_curve, auc
from tick.linear_model import LogisticRegression
from tick.dataset import fetch_tick_dataset
train_set = fetch_tick_dataset('binary/adult/adult.trn.bz2')
test_set = fetch_tick_dataset('binary/adult/adult.tst.bz2')
learner = LogisticRegression()
learner.fit(train_set[0], train_set[1])
predictions = learner.predict_proba(test_set[0])
fpr, tpr, _ = roc_curve(test_set[1], predictions[:, 1])
plt.figure(figsize=(6, 5))
plt.plot(fpr, tpr, lw=2)
plt.title("ROC curve on adult dataset (area = {:.2f})".format(auc(fpr, tpr)))
plt.ylabel("True Positive Rate")
plt.xlabel("False Positive Rate")
plt.show()
(Source code, png, hires.png, pdf)
3. Models¶
In tick a model class gives information about a statistical model.
Depending on the case, it gives first order information (loss, gradient) or
second order information (hessian norm evaluation).
In this module, a model corresponds to the choice of a loss function, that are
described below. The advantages of using one or another loss are explained in
the documentation of the classes themselves.
The following table lists the different losses implemented for now in this
module, its associated class and label type.
Model |
Type |
Label type |
Class |
|---|---|---|---|
Linear regression |
Regression |
Continuous |
|
Logistic regression |
Classification |
Binary |
|
Poisson regression (identity link) |
Count data |
Integer |
|
Poisson regression (exponential link) |
Count data |
Integer |
|
Hinge loss |
Classification |
Binary |
|
Quadratic hinge loss |
Classification |
Binary |
|
Smoothed hinge loss |
Classification |
Binary |
3.1. Description of the available models¶
On the following graph we represent the different losses available in tick for
supervised linear learning. Note that ModelHuber,
ModelEpsilonInsensitive,
ModelModifiedHuber and
ModelAbsoluteRegression are available
through the tick.robust module.
(Source code, png, hires.png, pdf)
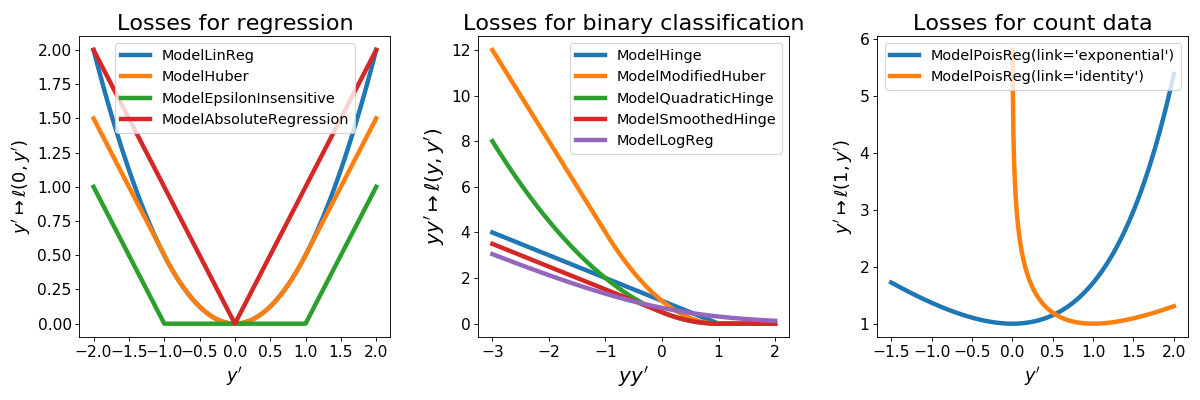
ModelLinReg¶
This is least-squares regression with loss
for \(y, y' \in \mathbb R\)
ModelLogReg¶
Logistic regression for binary classification with loss
for \(y \in \{ -1, 1\}\) and \(y' \in \mathbb R\)
ModelHinge¶
This is the hinge loss for binary classification given by
for \(y \in \{ -1, 1\}\) and \(y' \in \mathbb R\)
ModelQuadraticHinge¶
This is the quadratic hinge loss for binary classification given by
for \(y \in \{ -1, 1\}\) and \(y' \in \mathbb R\)
ModelSmoothedHinge¶
This is the smoothed hinge loss for binary classification given by
for \(y \in \{ -1, 1\}\) and \(y' \in \mathbb R\),
where \(\delta \in (0, 1)\) can be tuned using the smoothness parameter.
Note that \(\delta = 0\) corresponds to the hinge loss.
ModelPoisReg¶
Poisson regression with exponential link with loss corresponds to the loss
for \(y \in \mathbb N\) and \(y' \in \mathbb R\) and is obtained
using link='exponential'. Poisson regression with identity link, namely with loss
for \(y \in \mathbb N\) and \(y' > 0\) is obtained using
link='identity'.
3.2. The model class API¶
All model classes allow to compute the loss (value of the objective function \(f\)) and its gradient. Let us illustrate this with the logistic regression model. First, we need to simulate data.
import numpy as np
from tick.linear_model import SimuLogReg
from tick.simulation import weights_sparse_gauss
n_samples, n_features = 2000, 50
weights0 = weights_sparse_gauss(n_weights=n_features, nnz=10)
intercept0 = 1.
X, y = SimuLogReg(weights0, intercept=intercept0, seed=123,
n_samples=n_samples, verbose=False).simulate()
Now, we can create the model object for logistic regression
import numpy as np
from tick.linear_model import ModelLogReg
model = ModelLogReg(fit_intercept=True).fit(X, y)
print(model)
outputs
{
"dtype": "float64",
"fit_intercept": true,
"n_calls_grad": 0,
"n_calls_loss": 0,
"n_calls_loss_and_grad": 0,
"n_coeffs": 51,
"n_features": 50,
"n_passes_over_data": 0,
"n_samples": 2000,
"n_threads": 1,
"name": "ModelLogReg"
}
Printing any object in tick returns a json formatted description of it.
We see that this model uses 50 features, 51 coefficients (including the intercept),
and that it received 2000 sample points. Now we can compute the loss of the model using
the loss method (its objective, namely the value of the function \(f\)
to be minimized) by using
coeffs0 = np.concatenate([weights0, [intercept0]])
print(model.loss(coeffs0))
which outputs
0.3551082120992899
while
print(model.loss(np.ones(model.n_coeffs)))
outputs
5.793300908869233
which is explained by the fact that the loss is larger for a parameter which is far from
the ones used for the simulation.
The gradient of the model can be computed using the grad method
_, ax = plt.subplots(1, 2, sharey=True, figsize=(9, 3))
ax[0].stem(model.grad(coeffs0))
ax[0].set_title(r"$\nabla f(\mathrm{coeffs0})$", fontsize=16)
ax[1].stem(model.grad(np.ones(model.n_coeffs)))
ax[1].set_title(r"$\nabla f(\mathrm{coeffs1})$", fontsize=16)
which plots
(Source code, png, hires.png, pdf)
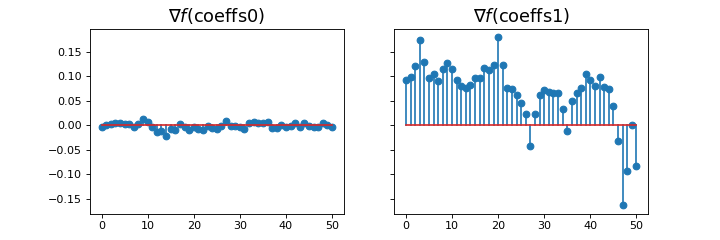
We observe that the gradient near the optimum is much smaller than far from it.
Model classes can be used with any solver class, by simply passing them through
the solver’s set_model method, see tick.solver.
What’s under the hood?
All model classes have a loss and grad method, that are used by batch
algorithms to fit the model. These classes contains a C++ object, that does the
computations. Some methods are hidden within this C++ object, and are accessible
only through C++ (such as loss_i and grad_i that compute the gradient
using the single data point \((x_i, y_i)\)). These hidden methods are used
in the stochastic solvers, and are available through C++ only for efficiency.
4. Simulation¶
Simulation of several linear models can be done using the following classes. All simulation classes simulates a features matrix \(\boldsymbol X\) with rows \(x_i\) and a labels vector \(y\) with coordinates \(y_i\) for \(i=1, \ldots, n\), that are i.i.d realizations of a random vector \(X\) and a scalar random variable \(Y\). The conditional distribution of \(Y | X\) is \(\mathbb P(Y=y | X=x)\), where \(\mathbb P\) depends on the considered model.
Model |
Distribution \(\mathbb P(Y=y | X=x)\) |
Class |
|---|---|---|
Linear regression |
\(\text{Normal}(w^\top x + b, \sigma^2)\) |
|
Logistic regression |
\(\text{Binomial}(w^\top x + b)\) |
|
Poisson regression (identity link) |
\(\text{Poisson}(w^\top x + b)\) |
|
Poisson regression (exponential link) |
\(\text{Poisson}(e^{w^\top x + b})\) |
|
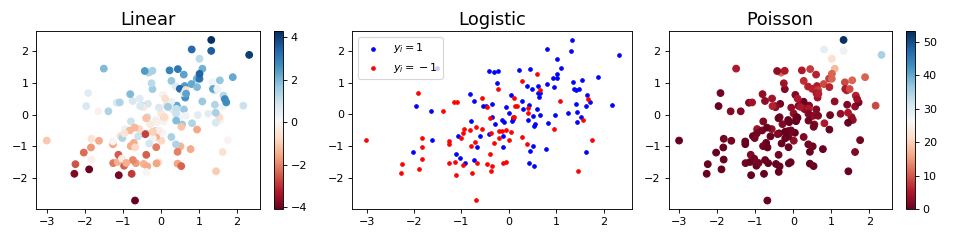
Example
"""
=============================
Linear models data simulation
=============================
Generates Linear, Logistic and Poisson regression realizations given a
weight vector.
"""
import matplotlib.pyplot as plt
import numpy as np
from tick.linear_model import SimuLinReg, SimuLogReg, SimuPoisReg
n_samples, n_features = 150, 2
weights0 = np.array([0.3, 1.2])
intercept0 = 0.5
simu_linreg = SimuLinReg(weights0, intercept0, n_samples=n_samples, seed=123,
verbose=False)
X_linreg, y_linreg = simu_linreg.simulate()
simu_logreg = SimuLogReg(weights0, intercept0, n_samples=n_samples, seed=123,
verbose=False)
X_logreg, y_logreg = simu_logreg.simulate()
simu_poisreg = SimuPoisReg(weights0, intercept0, n_samples=n_samples,
link='exponential', seed=123, verbose=False)
X_poisreg, y_poisreg = simu_poisreg.simulate()
plt.figure(figsize=(12, 3))
plt.subplot(1, 3, 1)
plt.scatter(*X_linreg.T, c=y_linreg, cmap='RdBu')
plt.colorbar()
plt.title('Linear', fontsize=16)
plt.subplot(1, 3, 2)
plt.scatter(*X_logreg[y_logreg == 1].T, color='b', s=10, label=r'$y_i=1$')
plt.scatter(*X_logreg[y_logreg == -1].T, color='r', s=10, label=r'$y_i=-1$')
plt.legend(loc='upper left')
plt.title('Logistic', fontsize=16)
plt.subplot(1, 3, 3)
plt.scatter(*X_poisreg.T, c=y_poisreg, cmap='RdBu')
plt.colorbar()
plt.title('Poisson', fontsize=16)
plt.tight_layout()
plt.show()
(Source code, png, hires.png, pdf)