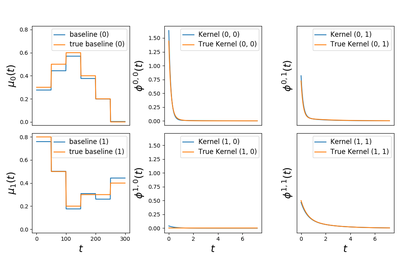
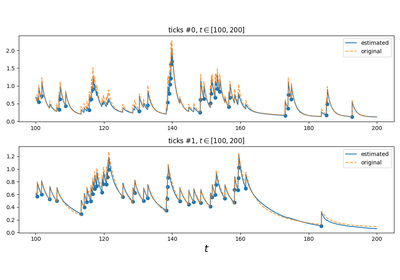
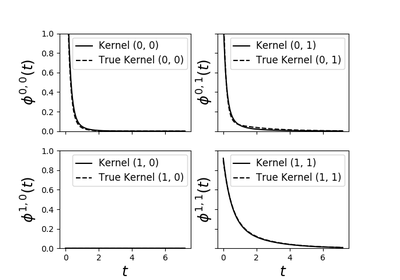
tick.hawkes.SimuHawkesSumExpKernels¶
-
class
tick.hawkes.SimuHawkesSumExpKernels(adjacency, decays, baseline=None, end_time=None, period_length=None, max_jumps=None, seed=None, verbose=True, force_simulation=False)[source]¶ Hawkes process with sum-exponential kernels simulation
They are defined by the intensity:
\[\forall i \in [1 \dots D], \quad \lambda_i(t) = \mu_i(t) + \sum_{j=1}^D \int \phi_{ij}(t - s) dN_j(s)\]where
\(D\) is the number of nodes
\(\mu_i(t)\) are the baseline intensities
\(\phi_{ij}\) are the kernels
\(dN_j\) are the processes differentiates
and with an exponential parametrisation of the kernels
\[\phi_{ij}(t) = \sum_{u=1}^{U} \alpha^u_{ij} \beta^u \exp (- \beta^u t) 1_{t > 0}\]where \(\alpha^u_{ij}\) are the intensities of the kernel and \(\beta^u\) its decays. The matrix of all \(\alpha\) is called adjacency matrix. Note that all nodes kernels share the same decays list.
- Parameters
baseline :
np.ndarrayorlistThe baseline of all intensities, also noted \(\mu(t)\). It might be three different types:
np.ndarray, shape=(n_nodes,) : One baseline per node is given. Hence baseline is assumed to be constant, ie. \(\mu_i(t) = \mu_i\)np.ndarray, shape=(n_nodes, n_intervals) :n_intervalsbaselines are given per node. This assumes parameterperiod_lengthis also given. In this case baseline is piecewise constant on intervals of sizeperiod_length / n_intervalsand periodic.listoftick.base.TimeFunction, shape=(n_nodes,) : One function is given per node, ie. \(\mu_i(t)\) is explicitely given.
adjacency :
np.ndarray, shape=(n_nodes, n_nodes, n_decays)Intensities of exponential kernels, also named \(\alpha^u_{ij}\)
decays :
np.ndarray, shape=(n_decays, )Decays of exponential kernels, also named \(\beta^u\) If a
floatis given, all decays are equal to this floatend_time :
float, default=NoneTime until which this point process will be simulated
max_jumps :
int, default=NoneSimulation will stop if this number of jumps in reached
seed :
int, default = NoneThe seed of the random sampling. If it is None then a random seed (different at each run) will be chosen.
force_simulation :
bool, default = FalseIf force is not set to True, simulation won’t be run if the matrix of the L1 norm of kernels has a spectral radius greater or equal to 1 as it would be unstable
- Attributes
timestamps :
listofnp.ndarray, size=n_nodesA list of n_nodes timestamps arrays, each array containing the timestamps of all the jumps for this node
n_decays :
intNumber of decays of the
HawkesSumExpKernel, also noted \(U\)simulation_time :
floatTime until which this point process has been simulated
n_total_jumps :
intTotal number of jumps simulated
tracked_intensity :
list[np.ndarray], size=n_nodesA record of the intensity with which this point process has been simulated. Note: you must call track_intensity before simulation to record it
intensity_tracked_times :
np.ndarrayThe times at which intensity has been recorded. Note: you must call track_intensity before simulation to record it
intensity_track_step :
floatStep with which the intensity has been recorded
-
__init__(adjacency, decays, baseline=None, end_time=None, period_length=None, max_jumps=None, seed=None, verbose=True, force_simulation=False)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
adjust_spectral_radius(spectral_radius)[source]¶ Adjust the spectral radius of the matrix of l1 norm of Hawkes kernels.
- Parameters
spectral_radius :
floatThe targeted spectral radius
-
get_baseline_values(i, t_values)¶ Outputs value of baseline depending on time
- Parameters
i :
intSelected dimension
t_values :
np.ndarrayValues baseline will be computed at
- Returns
output :
np.ndarrayValue of baseline
iatt_values
-
is_intensity_tracked()¶ Is intensity tracked thanks to track_intensity or not
-
mean_intensity()¶ Compute the mean intensity vector
-
reset()¶ Reset the process, so that is is ready for a brand new simulation
-
simulate()¶ Launch the simulation of data
-
spectral_radius()¶ Compute the spectral radius of the matrix of l1 norm of Hawkes kernels.
Notes
If the spectral radius is greater that 1, the hawkes process is not stable
-
threshold_negative_intensity(allow=True)¶ Threshold intensity to 0 if it becomes negative. This allows simulation with negative kernels
- Parameters
allow :
boolFlag to allow negative intensity thresholding
-
track_intensity(intensity_track_step=-1)¶ Activate the tracking of the intensity
- Parameters
intensity_track_step :
floatIf positive then the step the intensity vector is recorded every, otherwise, it is deactivated.
Notes
This method must be called before simulation